MYB (v-myb myeloblastosis viral oncogene homolog (avian))
2009-02-01 Liang Zhao , Diwakar R Pattabiraman , Thomas J Gonda AffiliationDiamantina Institute for Cancer, Immunology, Metabolic Medicine, University of Queensland, Australia
Identity
HGNC
LOCATION
6q23.3
LOCUSID
ALIAS
Cmyb,c-myb,c-myb_CDS,efg
FUSION GENES
DNA/RNA
Note
MYB was identified as the cellular homologue of v-myb, an oncogene found in two avian retroviruses (Avian Myeloblastosis Virus (AMV) and E26), which induce myeloblastic leukaemia and a mixed myeloid/erythroid leukaemia, respectively, in chickens.
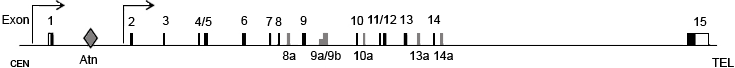
Description
The MYB gene spans 37.85 kb on the long arm of chromosome 6 and is transcribed in the centromere-to-telomere orientation.
Transcription
Multiple splicing variants of ~2.9 to 3.6 kb encode a series of MYB proteins with identical DNA binding domains but unique C-terminal domains. The best characterized MYB variant is p89 with additional exon 9B.
The first intron contains an elongation attenuation region (Atn) which is important in regulating transcription of MYB.
The first intron contains an elongation attenuation region (Atn) which is important in regulating transcription of MYB.
Proteins
Description
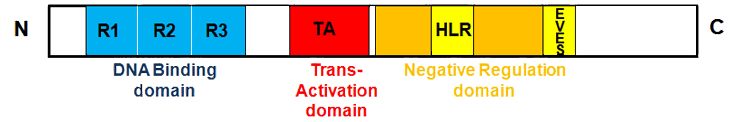
The MYB protein consists of three distinct functional domains: an N-terminal helix-turn-helix (HTH)-type DNA binding domain (DBD), a centrally located trans-activation domain (TAD) and a C-terminal negative regulatory domain (NRD).
The DBD consists of three tandem 52 amino acid repeat termed R1, R2 and R3, which are involved in recognition and binding to the consensus sequence PyAACT/GG, known as the MYB binding site (MBS). MYBs trans-activation of its target genes requires the TAD. The NRD negatively regulates the trans-activating and transforming capacity of MYB. Several motifs within the NRD have been identified, including a Heptad Leucine Repeat (HLR) and a highly conserved EVES motif. Disruption of the HLR motif results in enhancement of MYBs trans-activating and transforming capacities. The EVES motif is also involved in negative regulation of MYBs activity by mediating interactions between the N-terminus and the C-terminus of the MYB protein.
MYB protein is subject to post-translational modifications, including ubiquitination, sumoylation, acetylation and phosphorylation.
The DBD consists of three tandem 52 amino acid repeat termed R1, R2 and R3, which are involved in recognition and binding to the consensus sequence PyAACT/GG, known as the MYB binding site (MBS). MYBs trans-activation of its target genes requires the TAD. The NRD negatively regulates the trans-activating and transforming capacity of MYB. Several motifs within the NRD have been identified, including a Heptad Leucine Repeat (HLR) and a highly conserved EVES motif. Disruption of the HLR motif results in enhancement of MYBs trans-activating and transforming capacities. The EVES motif is also involved in negative regulation of MYBs activity by mediating interactions between the N-terminus and the C-terminus of the MYB protein.
MYB protein is subject to post-translational modifications, including ubiquitination, sumoylation, acetylation and phosphorylation.
Expression
MYB is expressed predominantly in immature progenitor cells of all haemopoietic lineages and these levels decrease as the cells progress towards terminal differentiation, although in differentiated T cells, MYB can be up-regulated upon activation.
Besides the haemopoietic system, MYB expression is also detected in colonic crypts and neurogenic niches.
MYB is highly expressed in almost all leukaemias. Overexpression of MYB is also detected in some solid tumors, such as breast cancer and colon cancer.
Besides the haemopoietic system, MYB expression is also detected in colonic crypts and neurogenic niches.
MYB is highly expressed in almost all leukaemias. Overexpression of MYB is also detected in some solid tumors, such as breast cancer and colon cancer.
Localisation
MYB is localized to the nucleus.
Function
MYB mostly operates as a transcriptional activator. It binds to its cognate binding site (MYB binding site MBS; consensus A/C A A C G/T G) on target genes and regulates their expression.
MYB is essential for the establishment of definitive haemopoiesis; as such, myb-/- mice die of anoxia by embryonic day 15. Conditional knockout mice have shown that myb is required at different stages of differentiation of the T and B lymphoid lineages. MYB is also reported to play a role in maintaining the haemopoietic stem cell pool. Interestingly, hypomorphic myb alleles result in an increase in platelet numbers in mouse models.
MYB is involved in maintaining the proliferation of progenitor cells. Knockdown of MYB in leukaemic cells and estrogen receptor positive breast cancer cells, where it is highly expressed, significantly slows down cell proliferation. Overexpressed or activated MYB suppresses normal differentiation and promotes leukaemic transformation. However, MYB also activates a number of haemopoietic-specific genes.
MYB is essential for the establishment of definitive haemopoiesis; as such, myb-/- mice die of anoxia by embryonic day 15. Conditional knockout mice have shown that myb is required at different stages of differentiation of the T and B lymphoid lineages. MYB is also reported to play a role in maintaining the haemopoietic stem cell pool. Interestingly, hypomorphic myb alleles result in an increase in platelet numbers in mouse models.
MYB is involved in maintaining the proliferation of progenitor cells. Knockdown of MYB in leukaemic cells and estrogen receptor positive breast cancer cells, where it is highly expressed, significantly slows down cell proliferation. Overexpressed or activated MYB suppresses normal differentiation and promotes leukaemic transformation. However, MYB also activates a number of haemopoietic-specific genes.
Homology
The MYB gene family contains the other two closely related members, MYBL1 (also known as A-MYB) and MYBL2 (also known as B-MYB).
The DBD is highly conserved between mammalian, chicken and drosophila MYB proteins, as well as MYBL1 and MYBL2. The MYB DBD also shares homology with other proteins, such as telomeric repeat binding factor -1, -2 (TRF1, TRF2) and cyclin D binding myb-like transcription factor 1 (DMTF1).
The DBD is highly conserved between mammalian, chicken and drosophila MYB proteins, as well as MYBL1 and MYBL2. The MYB DBD also shares homology with other proteins, such as telomeric repeat binding factor -1, -2 (TRF1, TRF2) and cyclin D binding myb-like transcription factor 1 (DMTF1).
Implicated in
Entity name
Disease
Translocation involving MYB and the T-cell receptor beta (TCRbeta) locus t(6;7)(q23;q34) and somatic genomic duplications at the C-MYB locus.
Cytogenetics
Molecular mapping of the chromosomal breakpoints show 2 discrete breakpoint clusters at 6q23.3: One located 5kb telomeric, 3 of the C-MYB gene, and the other 50kb more telomeric. Another gene (AHI1) was located in the vicinity of the t(6;7) breakpoints and was disrupted in some cases. In all cases, the translocation placed C-MYB in the vicinity of the TCRbeta regulatory sequence.
Oncogenesis
The abnormal regulation of C-MYB expression in this case confers a block of differentiation and continued proliferative capacity leading to its oncogenicity.
Entity name
Note
MYB is over-expressed in most human acute myeloid and lymphoid leukaemias. Several studies using antisense oligonucleotides and dominant negative forms of MYB have shown that MYB activity is essential for continued proliferation of AML and CML cells. Also, AML and CML cells are more sensitive to inhibition of MYB than their normal counterparts.
Entity name
AML that have MYST3-linked abnormalities
Disease
Genomic gain of the MYB locus.
Prognosis
MYST3-linked AMLs have been shown to be associated with poor prognosis.
Oncogenesis
A gain of the MYB locus at 6q23 was recurrently detected in AMLs that have MYST3-translocations. A consequent increase in MYB mRNA levels was also reported.
Entity name
Colorectal cancer
Prognosis
MYB is over-expressed in >80% of colorectal cancers. MYB expression correlates with poor prognosis for patients with colorectal cancer.
Oncogenesis
Robust mRNA expression and MYB mRNA amplifications were identified in colorectal cancer cell lines. Protein overexpression is a feature of these cell lines and primary cancers. Mutations in the first intron of MYB in the Atn region that regulates transcriptional elongation have been reported in some colorectal cancer cell lines and primary cancers.
Entity name
Breast cancers
Note
MYB over-expression.
Prognosis
Since Myb over-expression is closely related to estrogen receptor positivity in breast cancers, prognosis is generally positive. This is due to the less severe nature of ER+ breast tumours and the availability of established therapeutics.
Oncogenesis
64% of breast cancers express MYB, which also strongly correlates with ERalpha positivity.
Entity name
Breast cancer harboring BRCA1 mutations
Note
MYB amplification is seen in 29% of tumours in women with BRCA1 mutations.
Disease
MYB amplification.
Breakpoints
Article Bibliography
Other Information
Locus ID:
NCBI: 4602
MIM: 189990
HGNC: 7545
Ensembl: ENSG00000118513
Variants:
dbSNP: 4602
ClinVar: 4602
TCGA: ENSG00000118513
COSMIC: MYB
RNA/Proteins
Expression (GTEx)
Pathways
Protein levels (Protein atlas)
References
Citation
Liang Zhao ; Diwakar R Pattabiraman ; Thomas J Gonda
MYB (v-myb myeloblastosis viral oncogene homolog (avian))
Atlas Genet Cytogenet Oncol Haematol. 2009-02-01
Online version: http://atlasgeneticsoncology.org/gene/41466/myb-(v-myb-myeloblastosis-viral-oncogene-homolog-(avian))
