PHOX2B (paired-like homeobox 2b)
2013-04-01 Tiziana Bachetti , Isabella Ceccherini AffiliationUOC Medical Genetics, G Gaslini Institute, 16147 Genova, Italy
Identity
HGNC
LOCATION
4p13
IMAGE

LEGEND
Cytogenetic location of PHOX2B on chromosome 4.
LOCUSID
ALIAS
CCHS,NBLST2,NBPhox,PMX2B
DNA/RNA
Note
Genomic: NCBI Reference Sequence: NG_008243.1.

PHOX2B gene structure: untranslated regions (orange boxes), introns (black lines) and coding regions (blue boxes) are shown.
Description
The PHOX2B gene is 4888bp long; the coding region (CDS) is 945bp and is composed of 3 exons: exon 1 (241bp), exon 2 (188bp), exon 3 (516bp).
Other features: 5UTR: 361bp; 3UTR: 1725bp.
Other features: 5UTR: 361bp; 3UTR: 1725bp.
Transcription
mRNA: NCBI Reference Sequence NM_003924.3. The PHOX2B mRNA is 3218bp long; no alternative splice site is known.
Pseudogene
No pseudogene is reported.
Proteins
Note
NCBI Reference Sequence: NP_003915.2.
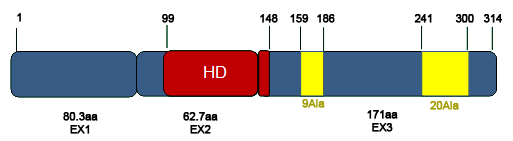
Representation of the PHOX2B protein. The three exons (blue) are shown, with the number of aminoacids (aa) they code for, in addition to the homeodomain (red) and the polyalanine stretches (yellow). In the upper part of the figure, the first and last aminoacid positions for each of these crucial regions is indicated.
Description
PHOX2B protein is 314 aminoacids long. A homeodomain region spanning exons 1 and 2 (from residue 99 to residue 148) is responsible for the binding of this transcription factor to target DNA elements. In exon 3 there are two sequences characterized by stretches of 9 and 20 alanine residues, encoded by GC(N) triplets. Their functional role is still unknown.
Expression
PHOX2B is expressed during neural development in central autonomic circuits and peripheral neural crest derivatives, and particularly in the retrotrapezoid nucleus, noradrenergic centres and hindbrain (Pattyn et al., 1997; Dubreuil et al., 2002; Stornetta et al., 2006; Kang et al., 2007).
PHOX2B expression is transcriptionally regulated by the PHOX2B protein itself, through its direct binding to four promoter elements which give rise to a positive autoregulatory loop (Cargnin et al., 2005). In addition, PHOX2B expression is known to be regulated at specific sympathetic and enteric nervous system developmental stages by E2a and Hand2 (Hashimoto et al., 2011). Finally, SOX10 (Nagashimada et al., 2012), as well as PHOX2A and HASH1 (reviewed by Brunet and Pattyn, 2002) have shown a degree of cross-regulation with respect to PHOX2B. Hoxb1 and Hoxb2 have also shown to form a trimer with Pbx1 and Meis 1 to regulate PHOX2B transcription (Samad et al., 2004).
PHOX2B expression is transcriptionally regulated by the PHOX2B protein itself, through its direct binding to four promoter elements which give rise to a positive autoregulatory loop (Cargnin et al., 2005). In addition, PHOX2B expression is known to be regulated at specific sympathetic and enteric nervous system developmental stages by E2a and Hand2 (Hashimoto et al., 2011). Finally, SOX10 (Nagashimada et al., 2012), as well as PHOX2A and HASH1 (reviewed by Brunet and Pattyn, 2002) have shown a degree of cross-regulation with respect to PHOX2B. Hoxb1 and Hoxb2 have also shown to form a trimer with Pbx1 and Meis 1 to regulate PHOX2B transcription (Samad et al., 2004).
Localisation
PHOX2B is localized within the nuclear compartment.
Function
PHOX2B is a transcription factor essential for the development of autonomic neural crest derivatives (Pattyn et al., 1999). It controls the development of motoneurons (Pattyn et al., 2000) and drives a somatic-to-visceral switch in cranial sensory pathways (DAutréaux et al., 2011).
PHOX2B regulates the transcriptional expression of several genes: TH (Lo et al., 1999), DBH (Adachi et al., 2000), PHOX2A (Flora et al., 2011), PHOX2B itself (Cargnin et al., 2005), RET (Bachetti et al., 2005a), TLX-2 (Borghini et al., 2006), ALK (Bachetti et al., 2010), SOX10 (Nagashimada et al., 2012), Hand1 (Vincentz et al., 2012), SCG2 (Wen et al., 2007), MSX1 (Revet et al., 2008).
CREB-binding protein (CREBBP/CBP) interacts with PHOX2B and serves as its coactivator to mediate synergistic trans-activation (Wu et al., 2009). Using a yeast two-hybrid screening, TRIM11 was isolated as an additional PHOX2B interacting protein (Hong et al., 2008).
PHOX2B regulates the transcriptional expression of several genes: TH (Lo et al., 1999), DBH (Adachi et al., 2000), PHOX2A (Flora et al., 2011), PHOX2B itself (Cargnin et al., 2005), RET (Bachetti et al., 2005a), TLX-2 (Borghini et al., 2006), ALK (Bachetti et al., 2010), SOX10 (Nagashimada et al., 2012), Hand1 (Vincentz et al., 2012), SCG2 (Wen et al., 2007), MSX1 (Revet et al., 2008).
CREB-binding protein (CREBBP/CBP) interacts with PHOX2B and serves as its coactivator to mediate synergistic trans-activation (Wu et al., 2009). Using a yeast two-hybrid screening, TRIM11 was isolated as an additional PHOX2B interacting protein (Hong et al., 2008).
Homology
The amino acid sequence of the human PHOX2B is 100% identical to those of the chimpanzee, rat and mouse, suggesting that the function of PHOX2B is highly conserved in Mammals.
Mutations
Note
There is a clear correlation between types of PHOX2B mutations and clinical manifestations. Indeed, while the vast majority of PHOX2B mutations identified in isolated Congenital Central Hypoventilation Syndrome (CCHS) are PARMs (Polyalanine repeats mutation), those present in HSCR-NB (Hirschsprungs disease-neuroblastoma) associated CCHS are non-PARMs (NPARM), namely either missense mutations or nucleotide deletions/insertions causing frameshifts of the open reading frame. Moreover, inherited and de novo missense and frameshift mutations in exons 2 and 3 of the PHOX2B gene have been detected in both sporadic (NB) and syndromic (NB+CCHS) cases (Weese-Mayer et al., 2010; Bachetti et al., 2005b), thus suggesting that PHOX2B may play a role in isolated NB pathogenesis.
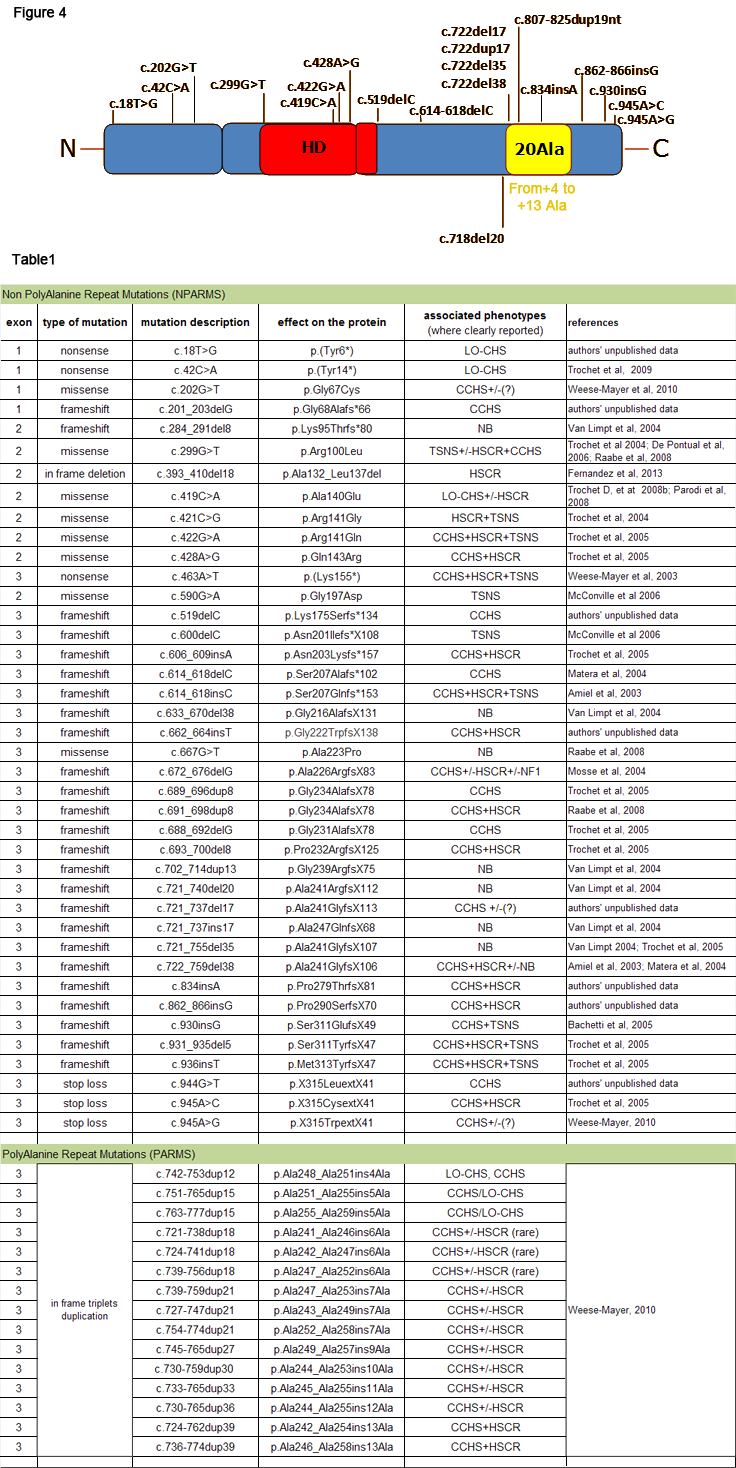
Fig.4: Localization of mutations within the PHOX2B gene. In the figure some PARMS (yellow) and NPARMS (black) PHOX2B mutations are shown (see also Weese-Mayer et al., 2010).
Table 1: NPARMS and PARMS mutations of PHOX2B in human disease. Table 1 reports PHOX2B mutations detected so far, with detailed description of the nucleotide and aminoacid changes. Associated phenotypes are also annotated. Legend: NB neuroblastoma, TSNS tumor sympathetic nervous system, CCHS congenital central hypoventilation syndrome, LO-CHS late onset central hypoventialtion syndrome, NF1 neurofibromatosis type 1, (?) undetermined associations.
Germinal
Germline PHOX2B mutations are responsible for Congenital Central Hypoventilation Syndrome (CCHS). The vast majority of mutations is represented by in frame triplet duplications of a sequence stretch coding for 20 Alanine residues in exon 3, also known as polyalanine (polyAla) expansions or PARM (Polyalanine repeats mutation). The duplication length is variable, starting from 12 bp up to 39 bp, thus leading from +4 Ala up to +13 Ala expansions (Amiel et al., 2003; Sasaki et al., 2003; Weese-Mayer et al., 2003; Matera et al., 2004; Trochet et al., 2008a).
Ninety percent of constitutive (germinal) mutations detected in CCHS patients are represented by polyAla expansions. Among these, 75% have arisen de novo while 25% is inherited from one parent (Bachetti et al., 2011; Meguro et al., 2012).
In addition, germline PHOX2B missense, frameshift, nonsense non polyAla mutations (NPARMs) have been detected in a small fraction of mainly syndromic patients characterized by CCHS+other neurocristophaties such as neuroblastoma (NB) and/or Hirschsprungs disease (HSCR) (Trochet et al., 2005; Matera et al., 2004). A few specific missense PHOX2B mutations have been detected in isolated NB and/or HSCR, as reported in Table 1.
In frame deletions within the polyAla stretch have been identified in healthy subjects, but also in association with schizophenia (Toyota et al., 2004).
Ninety percent of constitutive (germinal) mutations detected in CCHS patients are represented by polyAla expansions. Among these, 75% have arisen de novo while 25% is inherited from one parent (Bachetti et al., 2011; Meguro et al., 2012).
In addition, germline PHOX2B missense, frameshift, nonsense non polyAla mutations (NPARMs) have been detected in a small fraction of mainly syndromic patients characterized by CCHS+other neurocristophaties such as neuroblastoma (NB) and/or Hirschsprungs disease (HSCR) (Trochet et al., 2005; Matera et al., 2004). A few specific missense PHOX2B mutations have been detected in isolated NB and/or HSCR, as reported in Table 1.
In frame deletions within the polyAla stretch have been identified in healthy subjects, but also in association with schizophenia (Toyota et al., 2004).
Somatic
No CCHS patient has ever been proven to be a somatic mosaic for PHOX2B mutation; however, somatic and therefore germline mosaicism has been demonstrated in a proportion of CCHS parents (Parodi et al., 2008; Bachetti et al., 2011; Bachetti et al., 2013).
Somatic PHOX2B mutations have been identified in NB cell lines and NB tumor samples as reported in the Table 1.
Somatic PHOX2B mutations have been identified in NB cell lines and NB tumor samples as reported in the Table 1.
Implicated in
Entity name
PARMs (from +4 to +13 polyAla expansions)
Note
A correlation between the length of the expanded tract and the severity of the CCHS phenotype has already been reported (Matera et al., 2004; Weese-Mayer et al., 2010). Additional dysfunction of the autonomous nervous system, such as HSCR, NB, ocular defects, cardiac rythm alterations, etc., may be found in association with the especially largest polyAla expansions (Weese-Mayer et al., 2010).
Disease
Congenital Central Hypovention Syndrome (CCHS).
Prognosis
Congenital central hypoventilation syndrome (CCHS) is characterized by alveolar hypoventilation and autonomic dysregulation. It is a life-long, still untreatable disease.
Oncogenesis
Among PARMs, only subjects with the 20/29 and 20/33 genotypes have been identified to have tumors of neural crest origin (ganglioneuromas and ganglioneuroblastomas). No children with genotypes 20/24 to 20/28 have been identified with tumors of neural crest origin (Weese-Mayer et al., 2010).
Entity name
NPARMS: non polyAla repeat mutations, either germline or somatic (i.e. missense, nonsense, indels and loss of the stop codon mutations)
Note
These mutations, listed in Table 1, include missense, nonsense, loss of stop codon and small indels mutations.
Disease
Neuroblastoma and/or Hirschsprungs disease (either sporadic or in association with CCHS).
Entity name
Neuroblastoma
Note
In neuroblastoma cell lines and tumor samples, PHOX2B expression has turned out to be much higher than in normal tissues (Longo et al., 2008). Moreover, LOH in about 10% of the tumors and rare aberrant CpG dinucleotide methylation of 500bp of PHOX2B promoter region have been reported as NB associated molecular event (De Pontual et al., 2007).
Breakpoints
Note
Interestingly, other cytogenetic interstitial deletions, spanning from 0.29 to 2.6 Mb across the PHOX2B locus, have been detected in patients affected with atypical disorders, apparently sharing a loss of function/haploinsufficiency pathogenic mechanism, namely apparent-life threatening event including, Hirschsprung disease, transient neonatal hypoventilation and dysmorphic features. Neuroblastoma did not develop in any of these cases (Jennings et al., 2012).
Article Bibliography
Other Information
Locus ID:
NCBI: 8929
MIM: 603851
HGNC: 9143
Ensembl: ENSG00000109132
Variants:
dbSNP: 8929
ClinVar: 8929
TCGA: ENSG00000109132
COSMIC: PHOX2B
RNA/Proteins
Expression (GTEx)
Protein levels (Protein atlas)
References
Citation
Tiziana Bachetti ; Isabella Ceccherini
PHOX2B (paired-like homeobox 2b)
Atlas Genet Cytogenet Oncol Haematol. 2013-04-01
Online version: http://atlasgeneticsoncology.org/gene/126/phox2b-(paired-like-homeobox-2b)
